Choosing Threshold
The ccAFv2 classifier assigns a likelihood score to each cell cycle state, representing the confidence of the classification. To ensure reliable predictions, we can set a likelihood threshold, where:
-
Cells with a likelihood score above the threshold retain their predicted state.
-
Cells below the threshold are labeled as “Unknown” to prevent low-confidence classifications.
This method improves accuracy by removing ambiguous predictions, as demonstrated in our analysis of human neural stem cell (hNSCs) collected by Zeng et al., 2023.
Load packages
library(ccAFv2)
library(Seurat)
library(reticulate)
library(tensorflow)
Prepare Data
The U5 human neural stem cell (hNSC) dataset used to train the ccAFv2 is available for testing purposes here. This data has been QC’d and normalized using SCTransform.
We then apply the PredictCellCycle() function to classify cells into their respective cell cycle states using ccAFv2.
seurat_obj = readRDS('U5_normalized_ensembl.rds')
seurat_obj = PredictCellCycle(seurat_obj)
Selecting a Likelihood Threshold
To determine how varying likelihood thresholds affect classification, use ThresholdPlot(), which plots the relationship between likelihood thresholds, percentage of classified cells. This helps determine the optimal threshold for balancing classification confidence and data retention.
ThresholdPlot(seurat_obj)
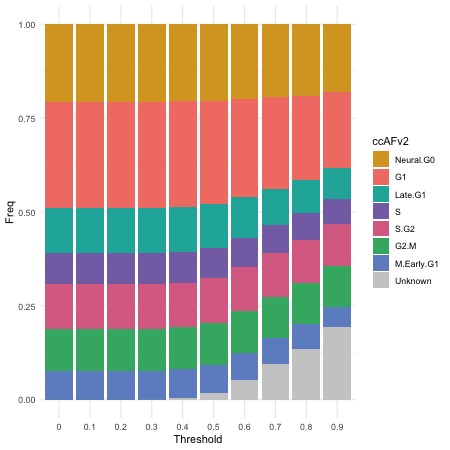
Based on our analysis, we set the likelihood threshold to 0.5. This ensures that only cells with a classification confidence of 50% or higher retain their predicted state, while others are labeled as “Unknown.”
AdjustCellCycleThreshold(seurat_obj, threshold = 0.5)