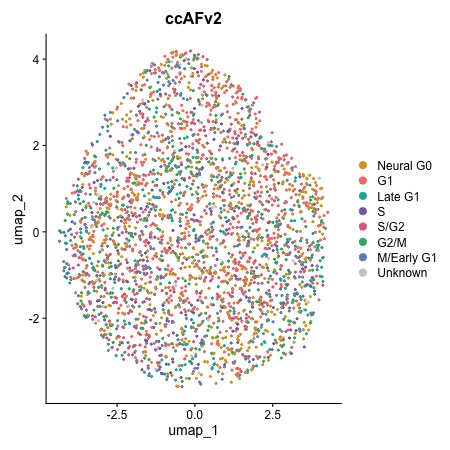
Cell Cycle Regression
After using ccAFv2 to predict the cell cycle, the cell cycle signal can be regressed out. Since the cell cycle significantly impacts gene expression, it is common practice to remove its effects and use the residual variance for further analysis. We support this using ccAFv2 marker genes, starting by calculating expression module scores for G0, Late G1, S, S/G2, G2/M, and M/Early G1 phases.
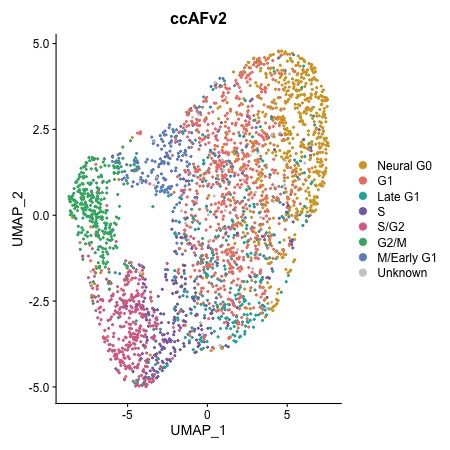
Before Regression
Prepare For Cell Cycle Regression
#Collect expression module scores for the cell cycle states
seurat_obj = PrepareForCellCycleRegression(seurat_obj)
#Regress these signatures out of the expression data
seurat_obj = SCTransform(seurat_obj, vars.to.regress = c("Neural.G0","Late.G1_exprs1", "S_exprs2", "S.G2_exprs3", "G2.M_exprs4", "M.Early.G1_exprs5"))
Removing the cell cycle from the U5 hNSCs leads to a random distribution because the cell cycle is the primary biological signal in this in vitro-grown cell line.
seurat_obj = RunPCA(seurat_obj)
seurat_obj = RunUMAP(seurat_obj, dims=1:10)
DimPlot.ccAFv2(seurat_obj)