About ccAFv2
Single-cell transcriptomics has revealed extensive cellular heterogeneity, with the cell cycle playing a key role. We developed a high-resolution cell cycle classifier, ccAFv2, using scRNA-seq data from human neural stem cells. ccAFv2 identifies six cell cycle states (G1, Late G1, S, S/G2, G2/M, and M/Early G1) and a quiescent-like G0 state, incorporating a tunable parameter to filter uncertain classifications. ccAFv2 generalizes well across cell types for S, S/G2, G2/M, and M/Early G1 states but shows reduced accuracy for G0, G1, and Late G1 in non-neuroepithelial cells. We applied ccAFv2 to classify cells, nuclei, and spatial transcriptomics data across species using various normalization methods and gene identifiers. The classifier also enables regression of cell cycle effects to uncover biological signals. 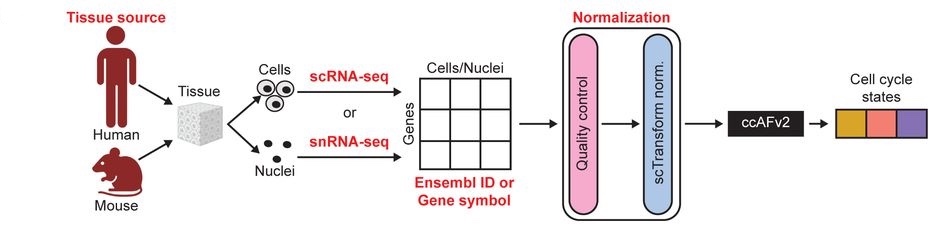
The ccAFv2 classifier builds on the foundation established by its predecessor (ccAFv1, O’Connor et al. 2021). ccAFv2 compares well against existing cell cycle state classifiers: ccAF (v1) (O’Connor et al. 2021), Seurat (Hao et al. 2021), Tricycle (Zheng et al. 2022), SchwabeCC (Schwabe et al. 2020; Zheng et al. 2022), reCAT (Liu et al. 2017), Peco (Hsiao et al. 2020) and Cyclone (Scialdone et al. 2015). 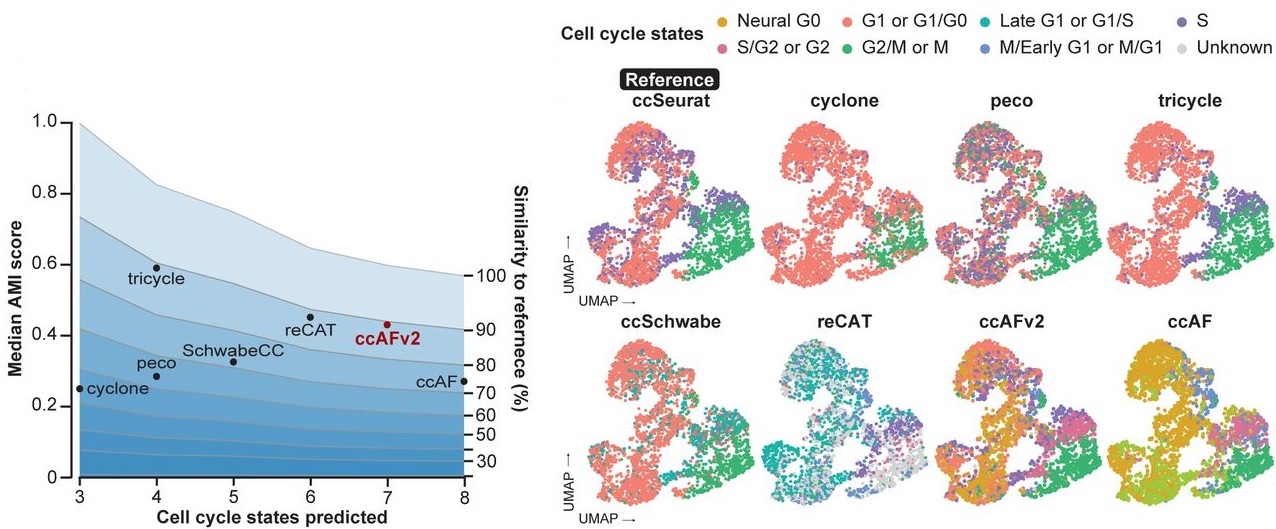
The core algorithm of the ccAFv2 is a fully connected artificial neural network (ANN) implemented using the Keras API (v2.12.0) that employs TensorFlow (v2.12.0) to construct ANNs. It takes the expression levels of 861 highly variable genes as input and classifies cells into seven distinct cell cycle states: Neural G0, G1, Late G1, S, S/G2, G2/M, and M/Early G1. The model incorporates dropout layers to prevent overfitting and uses Stochastic Gradient Descent (SGD) for optimization. The result is a powerful, flexible tool for analyzing cell cycle dynamics. 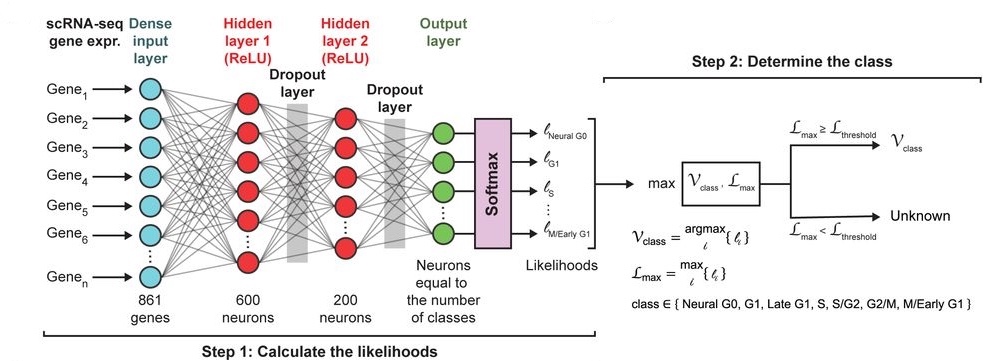
References
O’Connor SA, Feldman HM, Arora S, Hoellerbauer P, Toledo CM, Corrin P, Carter L, Kufeld M, Bolouri H, Basom R, et al. 2021. Neural G0: a quiescent-like state found in neuroepithelial-derived cells and glioma. Mol Syst Biol 17: e9522. PubMed
Hao Y, Hao S, Andersen-Nissen E, Mauck WM, Zheng S, Butler A, Lee MJ, Wilk AJ, Darby C, Zager M, et al. 2021. Integrated analysis of multimodal single-cell data. Cell 184: 3573–3587.e2 PubMed
Schwabe D, Formichetti S, Junker JP, Falcke M, Rajewsky N. 2020. The transcriptome dynamics of single cells during the cell cycle. Mol Syst Biol 16: e9946.PubMed
Zheng SC, Stein-O’Brien G, Augustin JJ, Slosberg J, Carosso GA, Winer B, Shin G, Bjornsson HT, Goff LA, Hansen KD. 2022. Universal prediction of cell-cycle position using transfer learning. Genome Biol 23: 41. PubMed
Liu Z, Lou H, Xie K, Wang H, Chen N, Aparicio OM, Zhang MQ, Jiang R, Chen T. 2017. Reconstructing cell cycle pseudo time-series via single-cell transcriptome data. Nat Commun 8: 22. PubMed
Hsiao CJ, Tung P, Blischak JD, Burnett JE, Barr KA, Dey KK, Stephens M, Gilad Y. 2020. Characterizing and inferring quantitative cell cycle phase in single-cell RNA-seq data analysis. Genome Res 30: 611–621. Full Text
Scialdone A, Natarajan KN, Saraiva LR, Proserpio V, Teichmann SA, Stegle O, Marioni JC, Buettner F. 2015. Computational assignment of cell-cycle stage from single-cell transcriptome data. Methods 85: 54–61 PubMed